INFERYS Rescoring
Boosting Peptide Identification

Unlocking the Intensity Dimension
Database search engines for bottom-up proteomics largely ignore peptide fragment ion intensities during the automated scoring of tandem mass spectra against protein databases. Using predicted fragment ion intensities to calculate additional scores helps to overcome this drawback. INFERYS® Rescoring for Sequest™ HT is an integrated workflow in Thermo Scientific™ Proteome Discoverer™ software, promising a deeper, more confident, and comprehensive proteomic data analysis. Classical search engine (SE) Sequest™ HT functions as a candidate generator. All candidate PSMs are predicted using INFERYS® and scored against experimental spectra using intensity-based figures of merit. Classical SE scores and intensity-based scores are aggregated by Percolator.

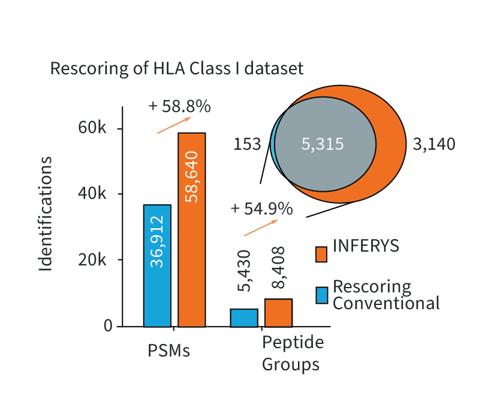
Rescoring Boosts Peptide Identifications
at 1% FDR
Rescoring fuses the well-established concept of database search with the advantages of state-of-the-art deep learning on your local PC. Rescoring provides merit for all sample types but demonstrates particular strength in challenging samples like immunopeptidomics experiments. Here, Rescoring enables better separating target and decoy identifications, thereby boosting the number of identified PSMs and peptides, while retaining the same false discovery rate.
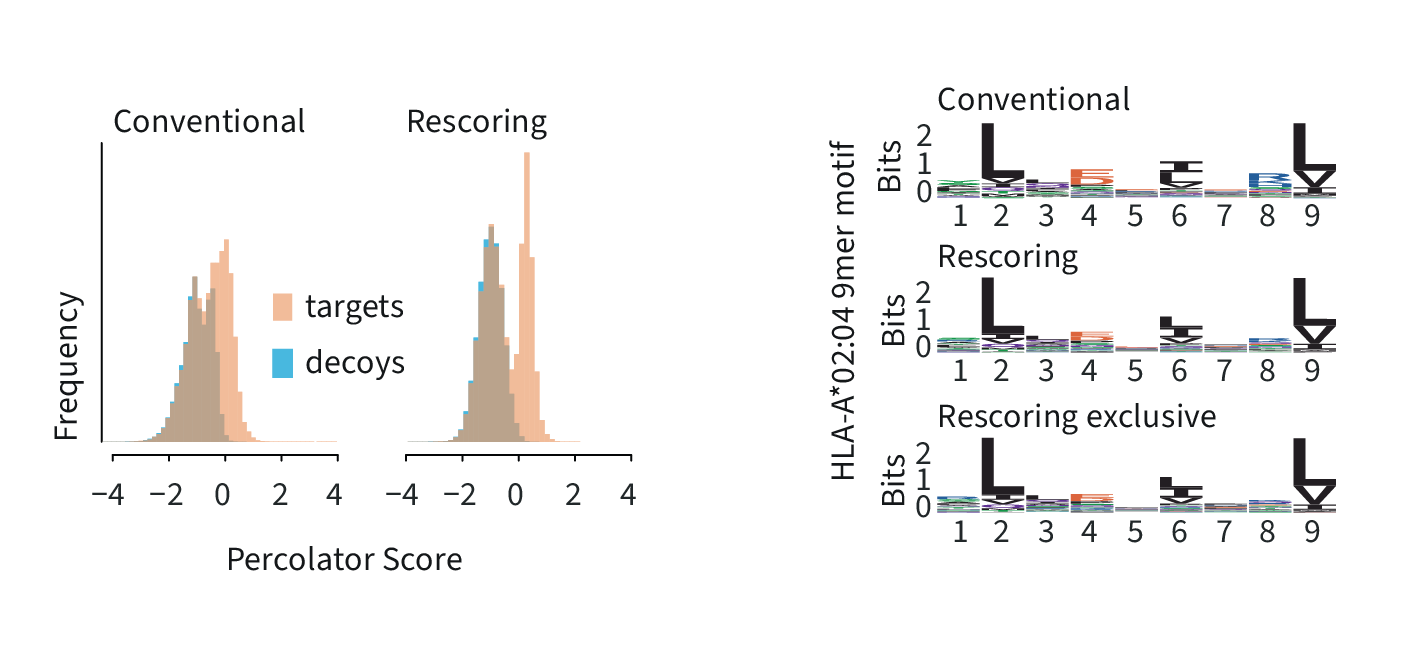
Number of peptide-spectrum-matches and peptide groups identified in immunopeptidomics data (Chong et al., Nat. Com., 2020) using different workflows. INFERYS Rescoring better separates targets and decoys, while retaining sequence motifs.
Left: Percolator SVM score distribution for the conventional and the INFERYS Rescoring workflow demonstrates better separation of target and decoy identifications, leading to a higher identification rate and better confidence.
Right: motif analysis for the HLA-A*02:04 allele comparing the identified peptides from both workflows and INFERYS Rescoring-exclusive identifications, resulting in a similar 9mer motif.
INFERYS Rescoring for Comprehensive Proteomic Data Processing
Testimonials
"INFERYS is a valuable new addition to the Proteome Discoverer (PD) environment. INFERYS can be utilized in multiple ways in PD, for the construction of deep learning-based spectral libraries as well as for rescuing MS/MS spectra that were incorrectly discarded as low quality by other tools in the PD pipeline"
Proteome Discoverer - A Community Enhanced Data Processing Suite for Protein Informatics. Proteomes. 2021 Mar 23;9(1):15. doi: https://doi.org/10.3390/proteomes9010015
"MS Amanda and SpectroMine can achieve comparable performance at sample loads exceeding 1 ng, but at sample loads below 1 ng, Sequest HT together with INFERYS results in the highest proteome coverage."
Ultrasensitive NanoLC-MS of Subnanogram Protein Samples Using Second Generation Micropillar Array LC Technology with Orbitrap Exploris 480 and FAIMS PRO. Anal Chem. 2021 Jun 29;93(25):8704-8710. doi: https://doi.org/10.1021/acs.analchem.1c00990
INFERYS Rescoring is integrated into Proteome Discoverer software. For more information and licensing, please visit thermofisher.com/proteomediscoverer. If you would like to learn what a customized deep learning model can do for your research, feel free to contact us!
Thermo Scientific™ Proteome Discoverer™ software is a trademark of Thermo Fisher Scientific, Inc. Sequest™ is a trademark of the University of Washington.
